library(tidyverse) # ggplot, lubridate, dplyr, stringr, readr...
library(praise)Solar Eclipse
The Data
This week we’re looking at the paths of solar eclipses in the United States. The data comes from NASA’s Scientific Visualization Studio.
eclipse_annular_2023 <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2024/2024-04-09/eclipse_annular_2023.csv')
eclipse_total_2024 <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2024/2024-04-09/eclipse_total_2024.csv')
eclipse_partial_2023 <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2024/2024-04-09/eclipse_partial_2023.csv')
eclipse_partial_2024 <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2024/2024-04-09/eclipse_partial_2024.csv')Putting the 2023 datasets together and the 2024 datasets together so that they can be plotted on the same map. The time variable represents the total duration of the eclipse: the amount of time from when the moon first comes in contact with the sun to when the moon is no longer in contact with the sun.
tot_23 <- eclipse_annular_2023 |>
mutate(time = eclipse_6 - eclipse_1,
eclipse = "total") |>
select(state, name, lat, lon, time, eclipse)
part_23 <- eclipse_partial_2023 |>
mutate(time = eclipse_5 - eclipse_1,
eclipse = "partial") |>
select(state, name, lat, lon, time, eclipse)
eclipse_23 <- rbind(tot_23, part_23) |>
mutate(time = as.numeric(time))tot_24 <- eclipse_total_2024 |>
mutate(time = eclipse_6 - eclipse_1,
eclipse = "total") |>
select(state, name, lat, lon, time, eclipse)
part_24 <- eclipse_partial_2024 |>
mutate(time = eclipse_5 - eclipse_1,
eclipse = "partial") |>
select(state, name, lat, lon, time, eclipse)
eclipse_24 <- rbind(tot_24, part_24) |>
mutate(time = as.numeric(time))library(ggnewscale)
states <- map_data("state")
ggplot(states) +
geom_polygon(fill = "white", color = "black",
aes(long, lat, group=group)) +
geom_point(data = filter(eclipse_23, lat < 51 & lat > 24 & lon < 0 & eclipse == "total"),
aes(x = lon, y = lat, color = time), size = .1) +
labs(x = "", y = "") +
scale_colour_gradientn(colors = c("blue", "turquoise")) +
labs(color = "time of overlap (sec)\nannular eclipse") +
new_scale_color() +
geom_point(data = filter(eclipse_23, lat < 51 & lat > 24 & lon < 0 & eclipse == "partial"),
aes(x = lon, y = lat, color = time), size = .001) +
scale_colour_gradientn(colors = c("red", "pink")) +
ggtitle("2023 annular solar eclipse") +
labs(color = "time of overlap (sec)\npartial eclipse")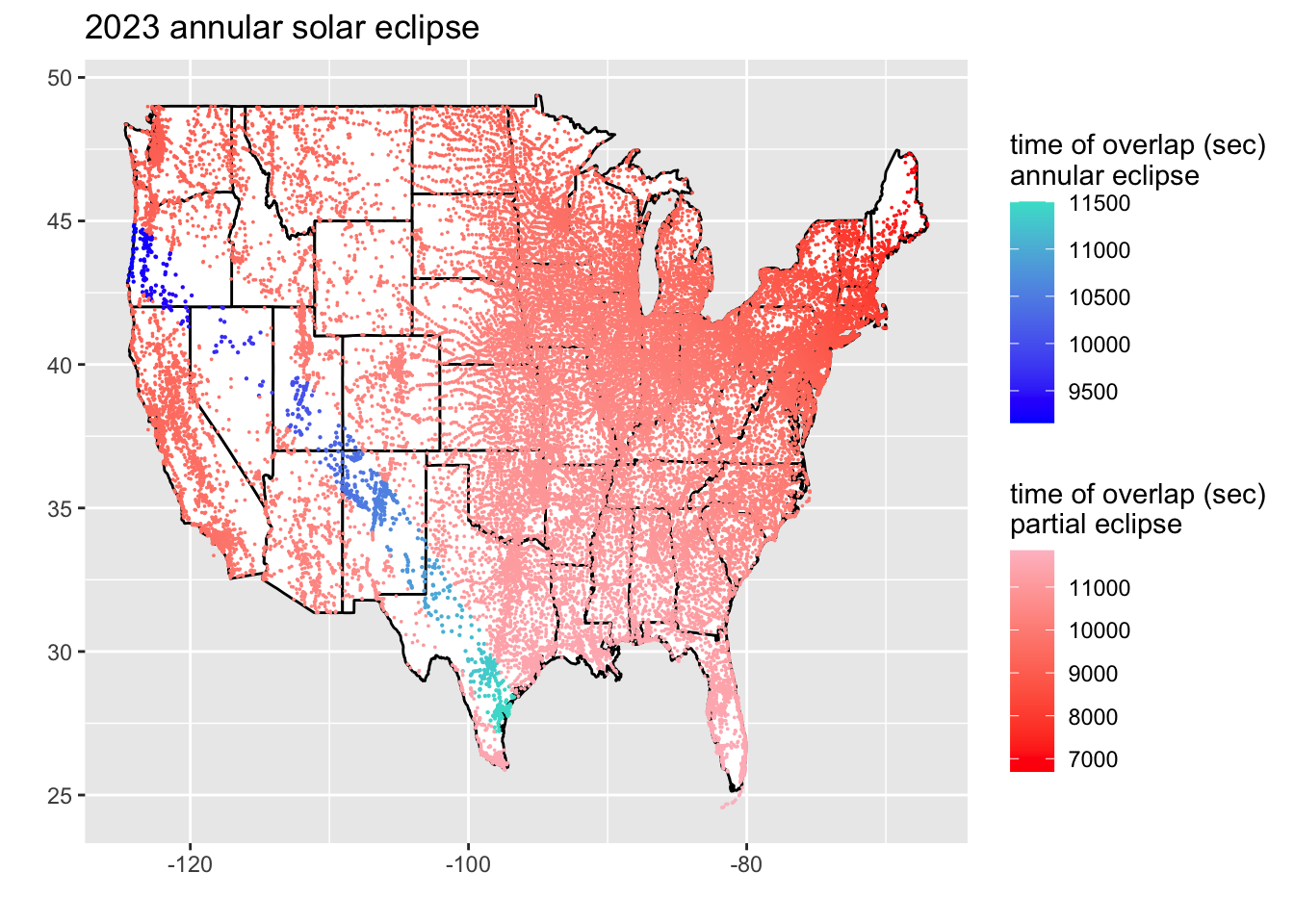
ggplot(states) +
geom_polygon(fill = "white", color = "black",
aes(long, lat, group=group)) +
geom_point(data = filter(eclipse_24, lat < 51 & lat > 24 & lon < 0 & eclipse == "total"),
aes(x = lon, y = lat, color = time), size = .0001) +
labs(x = "", y = "") +
scale_colour_gradientn(colors = c("blue", "turquoise")) +
labs(color = "time of overlap (sec)\ntotal eclipse") +
new_scale_color() +
geom_point(data = filter(eclipse_24, lat < 51 & lat > 24 & lon < 0 & eclipse == "partial"),
aes(x = lon, y = lat, color = time), size = .0001) +
scale_colour_gradientn(colors = c("red", "pink")) +
ggtitle("2024 total solar eclipse") +
labs(color = "time of overlap (sec)\npartial eclipse")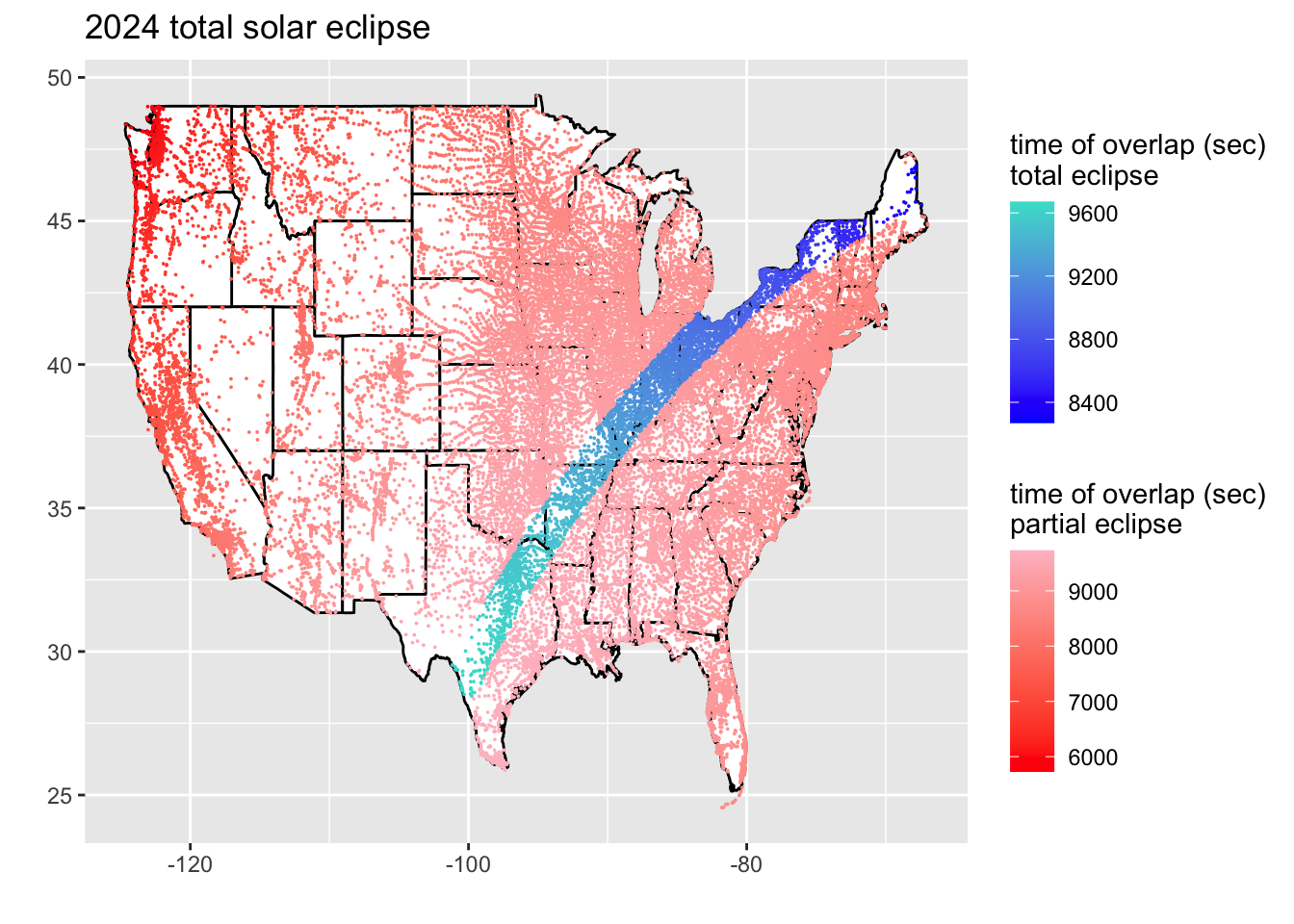
praise()[1] "You are superb!"